2 回答
TA贡献1871条经验 获得超13个赞
我对这个问题的解决方案是使用 NumPy 的快速傅里叶变换实现 numpy.fft.fft() 对数据应用离散傅里叶变换;这就是为什么数据在时间上均匀分布至关重要,因为 FFT 需要这样做。虽然 FFT 通常用于执行频谱分析,但所需的傅立叶系数与该函数的输出直接相关。
具体来说,该函数输出一系列i个复值系数c。傅立叶级数系数可使用以下关系式求得:
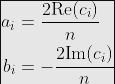
因此,FFT 允许直接计算傅立叶系数。这是我解决这个问题的 MWE,扩展了上面给出的示例:
import numpy as np
import matplotlib.pyplot as plt
# Set the number of equal-time bins to create.
n_bins = 101
# Set the number of Fourier coefficients to use.
n_coeff = 51
# Define a function to generate a Fourier series based on the coefficients determined by the Fast Fourier Transform.
# This also includes a series of phases x to pass through the function.
def create_fourier_series(x, coefficients):
# Begin the series with the zeroeth-order Fourier coefficient.
fourier_series = coefficients[0][0] / 2
# Now generate the first through n_coeff'th terms. The period is defined to be 1 since we're operating in phase
# space.
for n in range(1, n_coeff):
fourier_series += (fourier_coeff[n][0] * np.cos(2 * np.pi * n * x) + fourier_coeff[n][1] *
np.sin(2 * np.pi * n * x))
return fourier_series
# Create a dependent test variable to define the x-axis of the test data.
test_array = np.linspace(0, 1, n_bins) - 0.5
# Define some test data to try to apply a Fourier series to.
test_data = [0.9783883464566918, 0.979599093567252, 0.9821424606299206, 0.9857575507812502, 0.9899278899999995,
0.9941848228346452, 0.9978438300395263, 1.0003009205426352, 1.0012208923679058, 1.0017130521235522,
1.0021799664031628, 1.0027475606936413, 1.0034168260869563, 1.0040914266144825, 1.0047781181102355,
1.005520348837209, 1.0061899214145387, 1.006846206627681, 1.0074483048543692, 1.0078691461988312,
1.008318736328125, 1.008446947572815, 1.00862051262136, 1.0085134881422921, 1.008337095516569,
1.0079539881889774, 1.0074857334630352, 1.006747783037474, 1.005962048923679, 1.0049115434782612,
1.003812267822736, 1.0026427549407106, 1.001251963531669, 0.999898555335968, 0.9984976286266923,
0.996995982142858, 0.9955652088974847, 0.9941647321428578, 0.9927727076023389, 0.9914750532544377,
0.990212467710371, 0.9891098035363466, 0.9875998927875242, 0.9828093773946361, 0.9722532524271845,
0.9574084365384614, 0.9411012303149601, 0.9251820309477757, 0.9121488392156851, 0.9033119748549322,
0.9002445803921568, 0.9032760564202343, 0.91192435882353, 0.9249696964980555, 0.94071381372549,
0.957139088974855, 0.9721083392156871, 0.982955287937743, 0.9880613320235758, 0.9897455322896282,
0.9909590626223097, 0.9922601592233015, 0.9936513112840472, 0.9951442427184468, 0.9967071285988475,
0.9982921493123781, 0.9998775465116277, 1.001389230174081, 1.0029109110251453, 1.0044033691406251,
1.0057110841487276, 1.0069551867704276, 1.008118776264591, 1.0089884470588228, 1.0098663972602735,
1.0104514566473979, 1.0109849223300964, 1.0112043902912626, 1.0114717968750002, 1.0113343036750482,
1.0112205972495087, 1.0108811786407768, 1.010500276264591, 1.0099054552529192, 1.009353759223301,
1.008592596116505, 1.007887223091976, 1.0070715634615386, 1.0063525891472884, 1.0055587861271678,
1.0048733732809436, 1.0041832862669238, 1.0035913326848247, 1.0025318871595328, 1.000088536345776,
0.9963596140350871, 0.9918380684931506, 0.9873937281553398, 0.9833394624277463, 0.9803621496062999,
0.9786476100386117]
# Determine the fast Fourier transform for this test data.
fast_fourier_transform = np.fft.fft(test_data[n_bins / 2:] + test_data[:n_bins / 2])
# Create an empty list to hold the values of the Fourier coefficients.
fourier_coeff = []
# Loop through the FFT and pick out the a and b coefficients, which are the real and imaginary parts of the
# coefficients calculated by the FFT.
for n in range(0, n_coeff):
a = 2 * fast_fourier_transform[n].real / n_bins
b = -2 * fast_fourier_transform[n].imag / n_bins
fourier_coeff.append([a, b])
# Create the Fourier series approximating this data.
fourier_series = create_fourier_series(test_array, fourier_coeff)
# Create a figure to view the data.
fig, ax = plt.subplots(1, 1, figsize=(6, 6))
# Plot the data.
ax.scatter(test_array, test_data, color="k", s=1)
# Plot the Fourier series approximation.
ax.plot(test_array, fourier_series, color="b", lw=0.5)
输出如下:
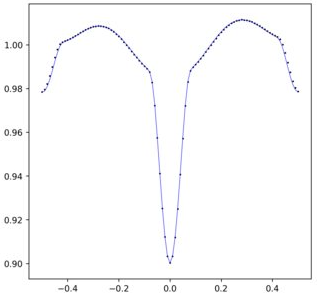
请注意,我如何定义 FFT(导入数据的后半部分,然后导入前半部分)是该数据生成方式的结果。具体来说,数据从 -0.5 到 0.5 运行,但 FFT 假设它从 0.0 运行到 1.0,因此需要进行此转换。
我发现这对于不包含非常尖锐和狭窄不连续性的数据非常有效。我很想听听是否有人对这个问题有其他建议的解决方案,我希望人们发现这个解释清晰且有帮助。
TA贡献1848条经验 获得超6个赞
不确定它是否对您有帮助;我写了一个程序来插入你的数据。这是使用 Buildingblocks==0.0.15 完成的
请看下面,
import matplotlib.pyplot as plt
from buildingblocks import bb
import numpy as np
Ydata = [0.9783883464566918, 0.979599093567252, 0.9821424606299206, 0.9857575507812502, 0.9899278899999995,
0.9941848228346452, 0.9978438300395263, 1.0003009205426352, 1.0012208923679058, 1.0017130521235522,
1.0021799664031628, 1.0027475606936413, 1.0034168260869563, 1.0040914266144825, 1.0047781181102355,
1.005520348837209, 1.0061899214145387, 1.006846206627681, 1.0074483048543692, 1.0078691461988312,
1.008318736328125, 1.008446947572815, 1.00862051262136, 1.0085134881422921, 1.008337095516569,
1.0079539881889774, 1.0074857334630352, 1.006747783037474, 1.005962048923679, 1.0049115434782612,
1.003812267822736, 1.0026427549407106, 1.001251963531669, 0.999898555335968, 0.9984976286266923,
0.996995982142858, 0.9955652088974847, 0.9941647321428578, 0.9927727076023389, 0.9914750532544377,
0.990212467710371, 0.9891098035363466, 0.9875998927875242, 0.9828093773946361, 0.9722532524271845,
0.9574084365384614, 0.9411012303149601, 0.9251820309477757, 0.9121488392156851, 0.9033119748549322,
0.9002445803921568, 0.9032760564202343, 0.91192435882353, 0.9249696964980555, 0.94071381372549,
0.957139088974855, 0.9721083392156871, 0.982955287937743, 0.9880613320235758, 0.9897455322896282,
0.9909590626223097, 0.9922601592233015, 0.9936513112840472, 0.9951442427184468, 0.9967071285988475,
0.9982921493123781, 0.9998775465116277, 1.001389230174081, 1.0029109110251453, 1.0044033691406251,
1.0057110841487276, 1.0069551867704276, 1.008118776264591, 1.0089884470588228, 1.0098663972602735,
1.0104514566473979, 1.0109849223300964, 1.0112043902912626, 1.0114717968750002, 1.0113343036750482,
1.0112205972495087, 1.0108811786407768, 1.010500276264591, 1.0099054552529192, 1.009353759223301,
1.008592596116505, 1.007887223091976, 1.0070715634615386, 1.0063525891472884, 1.0055587861271678,
1.0048733732809436, 1.0041832862669238, 1.0035913326848247, 1.0025318871595328, 1.000088536345776,
0.9963596140350871, 0.9918380684931506, 0.9873937281553398, 0.9833394624277463, 0.9803621496062999,
0.9786476100386117]
Xdata=list(range(0,len(Ydata)))
Xnew=list(np.linspace(0,len(Ydata),200))
Ynew=bb.interpolate(Xdata,Ydata,Xnew,40)
plt.figure()
plt.plot(Xdata,Ydata)
plt.plot(Xnew,Ynew,'*')
plt.legend(['Given Data', 'Interpolated Data'])
plt.show()
如果你想进一步编写代码,我也给出了代码,以便你可以查看源代码并学习:
import module
import inspect
src = inspect.getsource(module)
print(src)
添加回答
举报
