作者提供图片
利用知识图谱和AI来检索、筛选和总结医学文献应用程序和笔记本的配套代码可以在这里找到。在这里。
知识图谱(KGs)和大型语言模型(LLMs)是天作之合。我在之前的文章(点击这里)和文章(点击这里)中详细讨论了这两种技术的互补性。简而言之,LLMs的几个主要弱点包括黑盒模型和难以处理事实性知识,恰恰是KGs的优势所在。本质上,知识图谱就是一系列事实的集合,且完全透明可解释。
这篇文章主要介绍如何构建一个简单的图谱RAG应用。什么是RAG?RAG,即检索增强生成,是指检索相关资料来增强发给大语言模型(LLM)的提示,然后由LLM生成响应。图谱RAG是在检索部分使用知识图谱的RAG。如果你从未听说过图谱RAG,或想要复习一下,建议看看这个视频。
基本的想法是,与其将提示直接发送给未经过你数据训练的大型语言模型(LLM),你可以通过补充必要的相关信息来帮助LLM更准确地回答你的提示。比如,我可以将职位描述和我的简历复制到ChatGPT中,让它帮我写一封求职信。当我提供我的简历和申请职位的描述时,LLM能够更准确地回答我的提示:“写一封求职信”。由于知识图谱是设计来存储知识的,它们是存储内部数据并为LLM提示提供额外上下文信息的完美工具,这可以提高响应的准确性和上下文理解能力。
这项技术有多种多样的应用,例如客户服务机器人,药物 发现,生命科学领域的自动化监管报告生成,人力资源的招聘和管理,法律研究和写作,以及财富顾问助手。由于其广泛的适用性和提高大语言模型工具性能的潜力,图谱增强检索(RAG)迅速流行起来。以下是一张基于Google搜索兴趣变化的图表。
来源: https://trends.google.com/ (谷歌趋势页面)
Graph RAG 的搜索兴趣激增,甚至超过了像知识图谱和检索增强这样的术语。需要注意的是,Google Trends 测量的是 相对 搜索兴趣,而不是绝对的搜索次数。2024年7月,Graph RAG 的搜索兴趣激增,正好与微软在 宣布 GraphRAG 应用程序将在 GitHub 上发布的一周相吻合。
围绕Graph RAG的兴奋不仅仅局限于微软,而是更广泛。三星于2024年7月收购了知识图谱公司RDFox。宣布此次收购的这篇新闻文章并没有明确提到Graph RAG,但在福布斯网站上发表的这篇文章中提到,三星的一位发言人表示:“我们计划开发知识图谱技术,这是个性化AI的重要组成部分,并将其与生成式AI有机地结合起来,以支持特定用户的服务。”
2024年10月,领先的图数据库和语义网络公司Ontotext与知识图谱整理平台PoolParty的制造商合并,成立了Graphwise。根据官方新闻稿,此次合并旨在“推动图谱检索与生成(Graph RAG,一种基于图谱的检索与生成技术)这一类别的民主化”。
尽管围绕图RAG技术的部分讨论可能源自于对聊天机器人和生成式AI的更广泛的关注,但它确实反映了知识图谱在解决复杂现实世界问题方面的真正进步。一个例子是LinkedIn 应用了图RAG技术 来改善其客户服务技术支持。由于该工具能够检索相关的数据(如之前解决过的类似工单或问题)来为大语言模型(LLM)提供输入,因此回应更准确,平均解决时间从40小时缩短至15小时。
这篇帖子将介绍一个相对简单但很有说明性的例子,展示Graph RAG在实际中的工作方式。最终结果是一个非技术人员也能轻松使用的应用程序。和我在上一篇帖子中一样,我将使用来自PubMed的医学期刊文章数据集。这个应用程序旨在帮助医学领域的人员进行文献回顾。但同样的原则也可以应用于许多其他场景,这也是Graph RAG如此令人兴奋的原因。
这个应用的结构和这篇帖子如下:
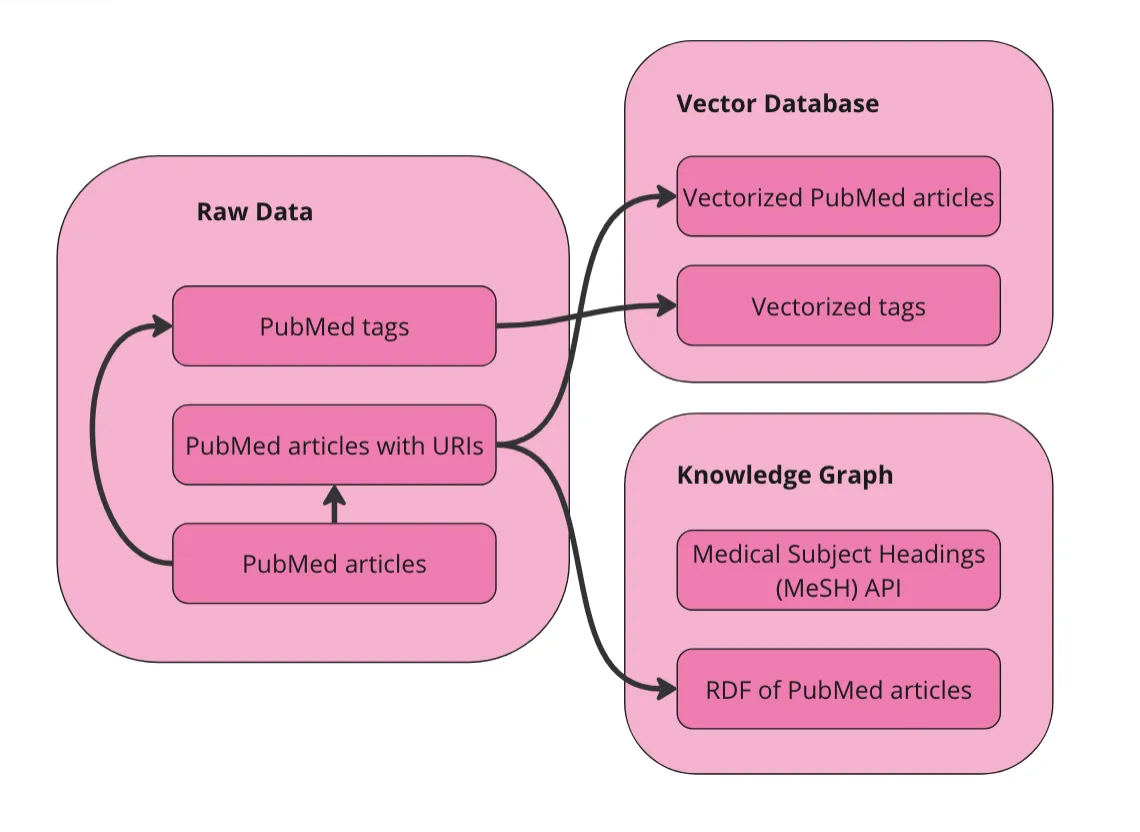
第一步是准备数据。我会在下面详细说明,但总体目标是,将原始数据向量化,并将其转换成RDF图。只要我们在向量化前保持URI与文章的关联,我们就可以在文章图和向量空间之间进行导航。这样我们就可以:
- 搜索文章: 利用向量数据库的力量,根据搜索词初步查找相关文章。我将使用向量相似度来检索与搜索词向量最相似的文章。
- 选择术语: 探索医学主题词表(MeSH)生物医学词汇表来选择用于筛选第1步中文章的术语。这个控制词汇表包含医学术语、别名、更具体化的概念以及其他许多属性和关系。
- 筛选和总结: 使用MeSH术语筛选文章以避免上下文污染。然后将筛选出的文章发送给大型语言模型,并附带一个额外的提示,如“用项目符号列出要点”。
在我们开始之前,先来了解一下关于这个应用和教程的几点说明:
- 此设置仅使用知识图谱来存储元数据,而不使用其他形式的元数据。之所以可以这样做,是因为我的数据集中的每篇文章都已经用一个丰富的受控词汇表中的术语进行了标记。我通过知识图谱构建结构和语义,并利用向量数据库进行基于相似度的检索,确保每项技术都用于其最擅长的领域。向量相似性可以表明“食道癌”与“口腔癌”在语义上的相似性,但知识图谱可以告诉我们“食道癌”和“口腔癌”之间的详细关系。
- 我为此应用程序使用的数据是来自PubMed的医学期刊文章集合(有关数据的更多信息请见下文)。我选择了这个数据集,因为它是结构化的,同时还包含每篇文章的摘要文本,并且这些文章已经用与一个公认的受控词汇表(MeSH)对齐的术语进行了标记。由于涉及的是医学文章,因此我将该应用程序命名为“医学图谱RAG”。但是,这种结构也能应用于其他领域,而不仅仅是医学领域。
- 我希望本教程和应用程序能够展示的是,通过在检索步骤中引入知识图谱,您可以提高RAG应用程序在准确性和可解释性方面的结果。我将展示知识图谱如何通过两种途径提高RAG应用程序的准确性:为用户提供一种过滤上下文的方法,以确保LLM仅接收最相关的信息;并利用领域专家维护和管理的领域特定受控词汇表来进行过滤。
- 本教程和应用程序并未直接展示知识图谱可以增强RAG应用程序的另外两种重要方式:治理、访问控制和合规性;以及效率和可扩展性。在治理方面,知识图谱不仅能通过过滤相关性来提高准确性,还能强制执行数据治理政策。例如,如果用户没有访问某一内容的权限,这些内容将不会出现在他们的RAG处理流程中。在效率和可扩展性方面,知识图谱能够打破数据孤岛,为构建、扩展和维护RAG应用程序提供坚实的基础。使用如MeSH这样的丰富受控词汇表来标记这些文章的元数据,可以确保此图谱RAG应用程序可以与其他系统集成,并降低其成为孤岛的风险。
准备数据的代码在下面这个笔记本里:点击这里这个笔记本里。
正如之前提到的,我再次决定使用来自PubMed库的这个数据集,此数据集包含50,000篇研究文章(授权CC0: 公共领域)。此数据集包含文章的标题和摘要,以及用于标签的元数据字段。这些标签来自医学主题词表(MeSH)控制词汇表。这些PubMed文章实际上只是关于文章的元数据——每篇文章都有摘要,但没有全文。数据以表格形式呈现,并已用MeSH术语标记。
我们可以直接将这个表格数据集向量化。我们也可以先将其转换为图(RDF)再进行向量化,但我没有这样做,而且我不确定这是否会对这种类型的数据最终结果有所帮助。向量化原始数据最重要的一点是,我们需要首先为每篇文章添加唯一资源标识符(URI)。URI是用于导航RDF数据的唯一标识符,对于我们在图中来回切换向量和实体来说是必要的。此外,我们将在向量数据库中为MeSH术语创建一个单独的集合。这将使用户可以无需了解这个受控词汇表就能搜索相关术语。以下是我们在准备数据时绘制的示意图。
作者的图片
我们在向量数据库中有两个可查询的集合:文章和术语。我们还以RDF格式将这些数据表示为图。由于MeSH有一个API,我将直接查询该API以获取术语的同义词和子概念。
在 Weaviate 中将数据向量化首先引入所需的包,然后配置 Weaviate 客户端。
导入 os
导入 json
导入 pandas 作为 pd
从 weaviate.util 导入 generate_uuid5
从 weaviate.classes.init 导入 Auth
client = weaviate.connect_to_weaviate_cloud(
cluster_url="XXX", # 请用你的 Weaviate Cloud URL 替换 XXX
auth_credentials=Auth.api_key("XXX"), # 请用你的 Weaviate Cloud key 替换 XXX
headers={'X-OpenAI-Api-key': "XXX"} # 请用你的 OpenAI API key 替换 XXX
)阅读 PubMed 上的期刊文章。我使用 Databricks 运行这个笔记本,所以你可能需要根据运行环境进行相应的调整。这里的目标只是将数据读取到 pandas 数据框中。
df = spark.sql("SELECT * FROM workspace.default.pub_med_multi_label_text_classification_dataset_processed").toPandas()如果你在本地运行,只需这样做就可以了。
df = pd.read_csv("PubMed Multi Label Text Classification Dataset Processed.csv") # 读取预处理过的PubMed多标签文本分类数据集然后稍微清理一下数据:
import numpy as np
# 将无穷大值替换为NaN,然后填充NaN值。
df.replace([np.inf, -np.inf], np.nan, inplace=True)
df.fillna('', inplace=True)
# 将列转换成字符串类型
df['Title'] = df['Title'].astype(str)
df['abstractText'] = df['abstractText'].astype(str)
df['meshMajor'] = df['meshMajor'].astype(str)现在我们需要为每篇文章创建一个唯一的统一资源标识符 (URI),并将其作为新的列添加。这是因为URI是连接文章的向量表示 (vector representation) 和知识图谱表示 (knowledge graph representation) 的关键。
import urllib.parse
from rdflib import Graph, RDF, RDFS, Namespace, URIRef, Literal
# 用于创建有效URI的函数
def create_valid_uri(base_uri, text):
if pd.isna(text):
return None
# 对文本进行编码以便在URI中使用
sanitized_text = urllib.parse.quote(text.strip().replace(' ', '_').replace('"', '').replace('<', '').replace('>', '').replace("'", "_"))
return URIRef(f"{base_uri}/{sanitized_text}")
# 用于创建文章的有效URI的函数
def create_article_uri(title, base_namespace="http://example.org/article/"):
"""
通过将非单词字符替换为下划线并进行URL编码来生成文章的URI。
参数:
title (str): 文章标题。
base_namespace (str): 文章URI的基本命名空间。
返回:
URIRef: 格式化的文章URI。
"""
if pd.isna(title):
return None
# 替换非单词字符为下划线
sanitized_title = re.sub(r'\W+', '_', title.strip())
# 压缩多个下划线为单个下划线
sanitized_title = re.sub(r'_+', '_', sanitized_title)
# URL编码术语
encoded_title = quote(sanitized_title)
# 将其与base_namespace连接,不添加额外的下划线
uri = f"{base_namespace}{encoded_title}"
return URIRef(uri)
# 向DataFrame添加一个新的文章URI列
df['Article_URI'] = df['Title'].apply(lambda title: create_valid_uri("http://example.org/article", title))我们也想创建一个包含所有用于标记文章的MeSH词的数据框。这将在稍后当我们需要查找相似的MeSH词时很有帮助。
# 用于清理和解析MeSH术语的函数
def parse_mesh_terms(mesh_list):
if pd.isna(mesh_list):
return []
return [
term.strip().replace(' ', '_')
for term in mesh_list.strip("[]'").split(',')
]
# 生成MeSH术语有效URI的函数
def create_valid_uri(base_uri, text):
if pd.isna(text):
return None
sanitized_text = urllib.parse.quote(
text.strip()
.replace(' ', '_')
.replace('"', '')
.replace('<', '')
.replace('>', '')
.replace("'", "_")
)
return f"{base_uri}/{sanitized_text}"
# 清理并解析所有MeSH术语
all_mesh_terms = []
for mesh_list in df["meshMajor"]:
all_mesh_terms.extend(parse_mesh_terms(mesh_list))
# 去重术语
unique_mesh_terms = list(set(all_mesh_terms))
# 创建一个包含MeSH术语及其URI的DataFrame
mesh_df = pd.DataFrame({
"meshTerm": unique_mesh_terms,
"URI": [create_valid_uri("http://example.org/mesh", term) for term in unique_mesh_terms]
})
# 打印DataFrame
print(mesh_df)将 articles 数据框中的数据向量化:
from weaviate.classes.config import Configure
# 定义这个集合
articles = client.collections.create(
name = "Article",
vectorizer_config=Configure.Vectorizer.text2vec_openai(), # 如果设置为 "none",你必须自己提供向量。也可以使用任何其他的 "text2vec-*"。
generative_config=Configure.Generative.openai(), # 确保使用 `generative-openai` 模块来执行生成查询
)
# 添加这些对象
articles = client.collections.get("Article")
with articles.batch.dynamic() as batch:
for index, 行 in df.iterrows():
batch.add_object({
"title": 行["标题"],
"abstractText": 行["摘要文本"],
"Article_URI": 行["文章URI"],
"meshMajor": 行["主要主题词"],
})现在将MeSH(医学主题词表)术语进行向量化处理:
#定义集合 terms
terms = client.collections.create(
name = "term",
vectorizer_config=Configure.Vectorizer.text2vec_openai(), # 如果设置为 "none",则必须自己提供向量。也可以是任何其他的 "text2vec-*"。
generative_config=Configure.Generative.openai(), # 确保使用 `generative-openai` 模块来进行生成查询
)
#添加对象到集合
terms = client.collections.get("term")
with terms.batch.dynamic() as batch:
for index, row in mesh_df.iterrows():
batch.add_object({ # 将 "meshTerm" 和 "URI" 添加到批次
"meshTerm": row["meshTerm"],
"URI": row["URI"],
})
``
你现在可以直接在向量化后的数据集上进行语义搜索、相似度搜索和RAG。这里我就不一一细说了,你可以查看我提供的[配套笔记本](https://github.com/SteveHedden/kg_llm/tree/main/graphRAGapp)中的代码,了解更多实现细节。
## 把数据变成知识图
我只是使用了我们在上次帖子([链接](https://medium.com/towards-data-science/how-to-implement-graph-rag-using-knowledge-graphs-and-vector-databases-60bb69a22759))中用过的相同代码来做这件事。我们基本上把数据中的每一行都转换成了知识图中的一个“文章”实体。然后我们给这些文章分配了标题、摘要和MeSH术语等属性。我们还将每个MeSH术语转换成一个实体。这段代码还会为每篇文章随机生成一个发布日期属性,并随机分配一个1到10之间的数字给名为访问量的属性。在这个演示中,我们不会用到这些属性。下面是从数据中生成的图的可视化表示。
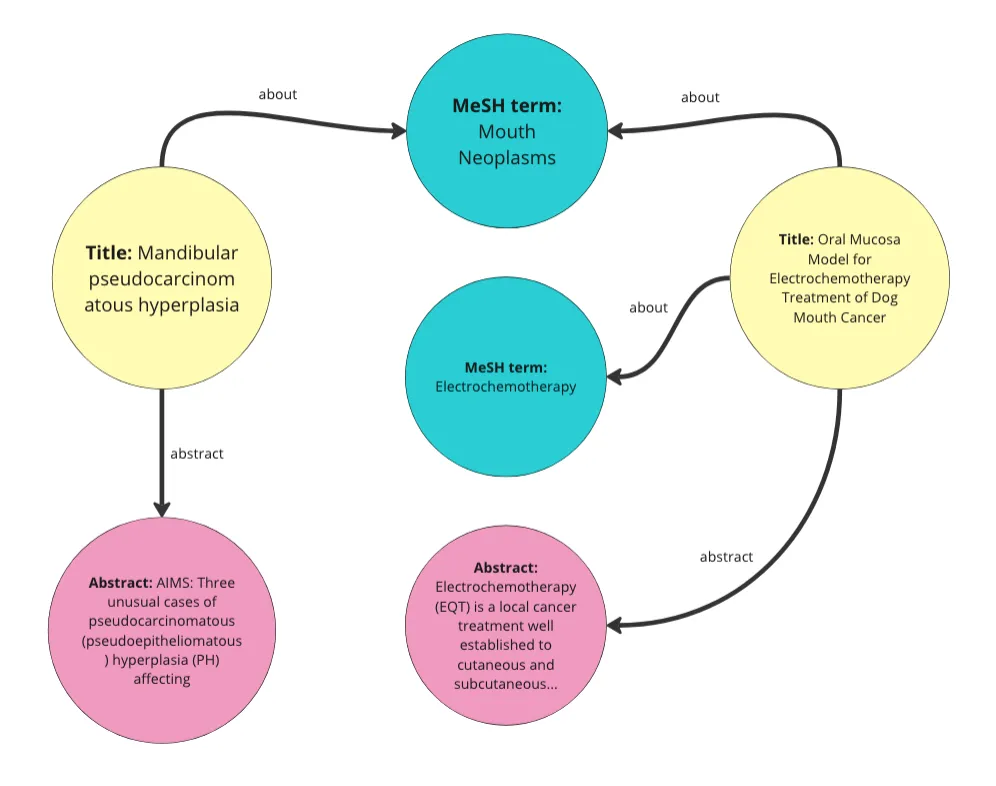
作者的图片
这是遍历数据框并将其转换为 RDF(资源描述框架)数据的方法:
from rdflib import Graph, RDF, RDFS, Namespace, URIRef, Literal
from rdflib.namespace import SKOS, XSD
import pandas as pd
import urllib.parse
import random
from datetime import datetime, timedelta
import re
from urllib.parse import quote
# --- 初始化 ---
g = Graph()
# 定义命名空间
schema = Namespace('http://schema.org/')
ex = Namespace('http://example.org/')
前缀 = {
'schema': schema,
'ex': ex,
'skos': SKOS,
'xsd': XSD
}
for p, ns in 前缀.items():
g.bind(p, ns)
# 定义类和属性
Article = URIRef(ex.Article)
MeSHTerm = URIRef(ex.MeSHTerm)
g.add((Article, RDF.type, RDFS.Class))
g.add((MeSHTerm, RDF.type, RDFS.Class))
title = URIRef(schema.name)
abstract = URIRef(schema.description)
date_published = URIRef(schema.datePublished)
access = URIRef(ex.access)
g.add((title, RDF.type, RDF.Property))
g.add((abstract, RDF.type, RDF.Property))
g.add((date_published, RDF.type, RDF.Property))
g.add((access, RDF.type, RDF.Property))
# 解析MeSH术语的函数
def parse_mesh_terms(mesh_list):
if pd.isna(mesh_list):
return []
return [term.strip() for term in mesh_list.strip("[]'").split(',')]
# 改进的转换函数
def convert_to_uri(term, base_namespace="http://example.org/mesh/"):
"""
通过替换空格和特殊字符为下划线,确保它以单个下划线开始和结束,并进行URL编码,来将MeSH术语转换为标准化的URI。
参数:
term (str): 要转换的MeSH术语。
base_namespace (str): URI的基础命名空间。
返回:
URIRef: 格式化的URI。
"""
if pd.isna(term):
return None
# 第一步:移除首尾的非单词字符(包括下划线)
stripped_term = re.sub(r'^\W+|\W+好的,现在我们有了数据的向量格式版本,以及数据的图(RDF)版本。每个向量都有关联的URI,该URI对应于KG中的一个实体,因此,我们可以在向量格式和图格式之间进行转换。
做一个应用我决定使用Streamlit来构建该图形RAG应用的界面。和上次一样,用户流程基本没变。
- 搜索文章: 首先,用户使用搜索词搜索文章。这完全依赖于向量数据库。用户的搜索词会被发送到向量数据库,然后返回与搜索词最接近的10篇文章。
- 细化术语: 其次,用户决定使用哪些MeSH术语来过滤返回的结果。由于我们也对MeSH术语进行了向量化,用户可以输入自然语言提示以获取最相关的MeSH术语,以帮助过滤结果。然后,用户可以扩展这些术语以查看它们的替代名称和更加具体的子概念。用户可以选择他们想要用于过滤条件的任意数量的这些术语。
- 过滤与总结: 第三,用户将所选术语应用为原始10篇期刊文章的过滤条件。我们可以通过这种方式进行筛选,因为PubMed文章带有MeSH标签。最后,用户可以输入额外的提示词,与筛选后的期刊文章一起发送给大语言模型 (LLM)。这便是RAG应用的生成步骤部分。
我们一步一步来吧。你可以在我的GitHub上查看完整的应用和代码,下面是我项目的结构。
-- app.py (驱动应用并按需调用其他功能的函数的Python文件)
-- query_functions (包含查询用Python文件的文件夹)
-- rdf_queries.py (含有RDF查询的Python文件)
-- weaviate_queries.py (含有Weaviate查询的Python文件)
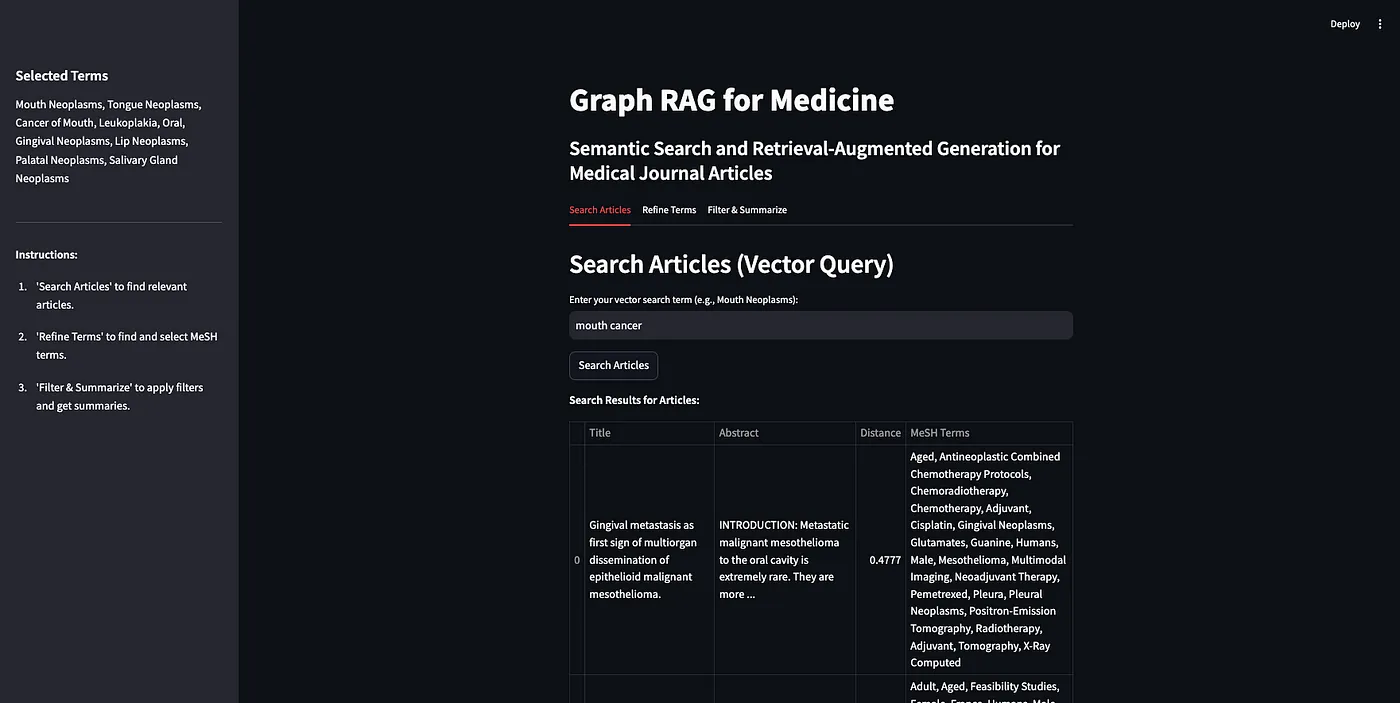
-- PubMedGraph.ttl (存储为RDF格式的PubMed数据)首先,我们要做的就是实现Weaviate的向量相似度搜索。由于我们已经将文章转化为向量,我们可以将搜索词发送到向量数据库,从而获得相似的文章。
图片由作者创作
在app.py中,查找相关期刊文章的主要功能是在向量检索库部分。
# --- 选项卡 1:搜索文章(向量查询)---
with tab_search:
st.header("搜索文章(向量查询)")
query_text = st.text_input("请输入您的向量查询词(例如,口腔肿瘤病):", key="vector_search")
if st.button("搜索文章", key="search_articles_btn"):
try:
client = initialize_weaviate_client()
article_results = query_weaviate_articles(client, query_text)
# 提取 URI
article_uris = [
result["properties"].get("article_URI")
for result in article_results
if result["properties"].get("article_URI")
]
# 将 article_uris 存储在 session 状态中
st.session_state.article_uris = article_uris
st.session_state.article_results = [
{
"标题": result["properties"].get("title", "未提供"),
"摘要": (result["properties"].get("abstractText", "未提供")[:100] + "..."),
"匹配度": result["distance"],
"MeSH 条目": ", ".join(
ast.literal_eval(result["properties"].get("meshMajor", "[]"))
if result["properties"].get("meshMajor") else []
),
}
for result in article_results
]
client.close()
except Exception as e:
st.error(f"文章搜索时发生错误:{e}")
if st.session_state.article_results:
st.write("**文章搜索结果:**")
st.table(st.session_state.article_results)
else:
st.write("暂时没有找到文章。")该功能利用存储在weaviate_queries中的查询来初始化Weaviate客户端(initialize_weaviate_client),并查询文章(query_weaviate_articles)。然后我们以表格形式展示返回的文章,包括摘要、距离(与搜索词的相关性)和它们所带的MeSH标签。
在 weaviate_queries.py 文件中,查询 Weaviate 的函数大致如下所示:
# 用于查询Weaviate中文章的函数
def query_weaviate_articles(client, query_text, limit=10):
# 执行文章集合的向量搜索
response = client.collections.get("Article").query.near_text(
query=query_text,
limit=limit,
return_metadata=MetadataQuery(distance=True)
)
# 解析响应结果
results = []
for obj in response.objects:
# 处理每个响应对象
results.append({
"uuid": obj.uuid,
"属性": obj.properties,
"距离": obj.metadata.distance,
})
return results正如你所看到的,我在这里只限制了前十个结果,但你可以调整这个限制。这只是通过Weaviate的向量相似度搜索来返回相关匹配。
应用程序的最终结果是这样的:
作者供图
作为一个演示,我将搜索术语“治疗口腔癌症”。正如你看到的,返回了10篇文章之多,大部分都有关。这展示了向量检索的长处和短处。
我们可以通过最小的努力在数据上构建语义搜索功能。如上所示,我们所做的只是设置客户端并将数据发送到向量数据库。一旦我们的数据被向量化,我们就可以进行语义搜索、相似性搜索,甚至可以通过RAG(检索增强生成)来增强功能。我已经在配套的笔记本中包含了一些示例,但是更多细节可以在Weaviate的官方文档中找到。
如上所述,基于向量的检索方法的弱点在于它们是黑盒,难以处理事实性信息。在我们的例子中,看起来大多数文章都是关于某种癌症的治疗或疗法。有些文章专门讨论口腔癌,有些则讨论口腔癌的亚型,例如牙龈癌(牙龈癌)和硬腭癌(硬腭癌)。但也有一些文章讨论鼻咽癌(上咽喉癌)、下颌癌(下颌骨癌)和食道癌(食管癌)。然而,这些(上咽喉、下颌骨和食管)都不被认为是口腔癌。可以理解为什么一篇关于鼻咽部肿瘤放射疗法的文章会被认为与“口腔癌的治疗方法”相关,但如果只寻找口腔癌的治疗方法,这可能并不相关。如果我们直接将这十篇文章输入到大语言模型的提示中,让它总结不同的治疗选项,我们可能会收到错误的信息。
RAG 的目的是给大型语言模型提供一些特定的额外信息,以便更好地回答你的问题。如果这些信息不正确或不相关,可能会导致大型语言模型给出误导性的回答。这种情况常被称为“上下文中毒”。特别危险的是,尽管回答并不一定是事实错误(大型语言模型可能准确地总结了我们提供的信息),也不是基于不准确的数据(假设期刊文章本身是准确的),但它用错了信息来回答你的问题。在这种情况中,用户可能会获得如何治疗错误类型癌症的信息,这听起来非常糟糕。
优化术语知识图可以帮助提高回复的准确性,并通过优化来自向量数据库的结果来减少上下文污染的风险。下一步是选择我们要使用的MeSH术语来过滤文章。首先,我们对向量数据库中的Terms集合进行另一次向量相似度搜索。这是因为用户可能不熟悉MeSH术语表。在之前的例子中,我搜索了“关于治疗口腔癌症的”,但“口腔癌症”并不是MeSH中的术语——它们使用的是“口腔肿瘤”。我们希望用户能够无需预先了解这些术语就开始探索MeSH术语——无论采用哪种元数据进行内容标记,这都是一个良好的做法。
作者供图
获取相关MeSH术语的功能与之前的Weaviate查询非常相似。只需要将Article替换为term。
# 查询Weaviate获取MeSH术语的函数
def query_weaviate_terms(client, query_text, limit=10):
# 在MeshTerm集合上进行向量搜索
response = client.collections.get("term").query.near_text(
query=query_text,
limit=limit,
return_metadata=MetadataQuery(distance=True)
)
# 解析响应内容
results = []
for obj in response.objects:
results.append({
"uuid": obj.uuid,
"properties": obj.properties,
"相似度": obj.metadata.distance,
})
return results这里就是应用的样子:
作者供图
正如你所见,我搜索了“口腔癌”,返回了最相关的术语。由于“口腔癌”并非MeSH词表中的术语,因此没有直接返回,不过“口腔肿瘤”却出现在了列表中。
下一步是让用户能够展开返回的术语,查看其替代名称和更具体的子概念。这需要查询MeSH API。这是应用程序中最棘手的部分,原因众多。最大的问题是Streamlit规定所有内容都有一个唯一的ID,而MeSH术语可能重复——如果其中一个术语是另一个术语的子项,那么当你展开父项时,子项就会重复。我认为我已经解决了大部分主要问题,应用程序应该可以正常运作了,但在这个阶段可能还会有需要发现的bug。
我们用到的功能在rdf_queries.py这个文件里。我们需要一个功能来获取术语的别名或同义词。
# 获取MeSH术语的概念三元组及其替代名称
def get_concept_triples_for_term(term):
term = sanitize_term(term) # 清理输入的术语(term)
sparql = SPARQLWrapper("https://id.nlm.nih.gov/mesh/sparql")
query = f"""
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX meshv: <http://id.nlm.nih.gov/mesh/vocab#>
PREFIX mesh: <http://id.nlm.nih.gov/mesh/>
SELECT ?subject ?p ?pLabel ?o ?oLabel
FROM <http://id.nlm.nih.gov/mesh>
WHERE {{
?subject rdfs:label "{term}"@en .
?subject ?p ?o .
FILTER(CONTAINS(STR(?p), "concept"))
OPTIONAL {{ ?p rdfs:label ?pLabel . }}
OPTIONAL {{ ?o rdfs:label ?oLabel . }}
}}
"""
try:
sparql.setQuery(query)
sparql.setReturnFormat(JSON)
results = sparql.query().convert()
triples = set()
for result in results["results"]["bindings"]:
obj_label = result.get("oLabel", {}).get("value", "No label")
triples.add(sanitize_term(obj_label)) # 清理术语后再添加
# 添加清理后的术语本身,确保其包含在结果中
triples.add(sanitize_term(term))
return list(triples)
except Exception as e:
print(f"在获取术语 '{term}' 的概念三元组时出错:{e}")
return []我们也需要一些功能来获取给定术语的所有更具体的(子)术语。我有两个函数可以做到这一点——一个用来获取术语的直接子项,另一个是递归函数,可以返回给定深度的所有子项。
# 获取MeSH术语的更具体术语
def get_narrower_concepts_for_term(term):
term = sanitize_term(term) # 清理输入术语
sparql = SPARQLWrapper("https://id.nlm.nih.gov/mesh/sparql")
query = f"""
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX meshv: <http://id.nlm.nih.gov/mesh/vocab#>
PREFIX mesh: <http://id.nlm.nih.gov/mesh/>
SELECT ?narrowerConcept ?narrowerConceptLabel
WHERE {{
?broaderConcept rdfs:label "{term}"@en .
?narrowerConcept meshv:broaderDescriptor ?broaderConcept .
?narrowerConcept rdfs:label ?narrowerConceptLabel .
}}
"""
try:
sparql.setQuery(query)
sparql.setReturnFormat(JSON)
results = sparql.query().convert()
concepts = set()
for result in results["results"]["bindings"]:
subject_label = result.get("narrowerConceptLabel", {}).get("value", "无标签")
concepts.add(sanitize_term(subject_label)) # 清理术语后再添加
return list(concepts)
except Exception as e:
print(f"获取术语 '{term}' 的更具体术语时出错: {e}")
return []
# 递归获取所有更具体术语
def get_all_narrower_concepts(term, depth=2, current_depth=1):
term = sanitize_term(term) # 清理输入术语
all_concepts = {}
try:
narrower_concepts = get_narrower_concepts_for_term(term)
all_concepts[sanitize_term(term)] = narrower_concepts
if current_depth < depth:
for concept in narrower_concepts:
child_concepts = get_all_narrower_concepts(concept, depth, current_depth + 1)
将子术语合并到all_concepts中
except Exception as e:
print(f"获取术语 '{term}' 的所有更具体术语时出错: {e}")
返回所有术语步骤2的另一个重要部分是让用户选术语加到“已选术语”列表中。这些会出现在屏幕左边的侧边栏里。这一步还有很多改进的空间,例如:
- 无法一次性清除所有内容,但可以清除缓存或在需要时刷新浏览器。
- 没有“选择所有更具体的子概念”的选项,这会很有帮助。
- 没有添加过滤规则的选项。目前,我们假设文章必须包含术语A、术语B、术语C等,例如。最终排名是根据文章中被标记的术语数量来确定的。
下面来看看这个应用的样貌吧
图片由作者创作。
我可以展开“口腔肿瘤”来查看其替代名称,在这种情况下是“这里的口腔癌症”,以及所有更详细的子概念。就像你看到的那样,大多数更详细的子概念都有自己的子概念,你也可以展开它们。为了演示的目的,我将选择所有与口腔肿瘤相关的子概念。
作者的图片
这一步非常重要,不仅因为它允许用户筛选搜索结果,还因为它让用户自己探索并学习MeSH图。比如,用户可以在这里发现鼻咽肿瘤并不是口腔肿瘤的一部分。
筛选和:总结现在你已经有了文章和筛选词,你可以应用筛选并总结结果。这是我们将第一步中返回的原始10篇文章与精炼后的MeSH术语列表相结合的地方。我们允许用户在将提示发送给LLM之前添加更多背景信息。
图片由作者创作
我们这样过滤:首先,我们需要从最初的搜索中获取这10篇文章的链接。接着,我们可以在知识图谱中查找哪些文章被标上了相关的MeSH术语。同时,我们还会保存这些文章的摘要,供下一步使用。这里可以基于访问控制或其他用户设定的参数(例如作者、文件类型、发表日期等)进行过滤。虽然我在这个应用中没有加入这些功能,但我确实添加了访问控制和发表日期的选项,以防将来在UI中加入这些功能。
这里就是app.py中的代码:
if st.button("筛选文章"):
try:
# 检查是否从第一个选项卡获取了URIs
if "article_uris" in st.session_state and st.session_state.article_uris:
article_uris = st.session_state.article_uris
# 将URI列表转换为VALUES子句或FILTER中的字符串
article_uris_string = ", ".join([f"<{str(uri)}>" for uri in article_uris])
SPARQL_QUERY = """
PREFIX schema: <http://schema.org/>
PREFIX ex: <http://example.org/>
SELECT ?article ?title ?abstract ?datePublished ?access ?meshTerm
WHERE {{
?article a ex:Article ;
schema:name ?title ;
schema:description ?abstract ;
schema:datePublished ?datePublished ;
ex:access ?access ;
schema:about ?meshTerm .
?meshTerm a ex:MeSHTerm .
FILTER (?article IN ({article_uris}))
}}
"""
# 将文章的URI插入查询中
query = SPARQL_QUERY.format(article_uris=article_uris_string)
else:
st.write("未从第一个选项卡选择文章。")
return
# 查询RDF并将结果保存在会话状态中
top_articles = query_rdf(LOCAL_FILE_PATH, query, final_terms)
st.session_state.filtered_articles = top_articles
if top_articles:
# 合并顶级文章的摘要并保存在会话状态中
def combine_abstracts(ranked_articles):
combined_text = " ".join(
[f"标题: {data['title']} 摘要: {data['abstract']}" for article_uri, data in
ranked_articles]
)
return combined_text
st.session_state.combined_text = combine_abstracts(top_articles)
else:
st.write("未找到与所选术语相关的文章。")
except Exception as e:
st.error(f"筛选文章时出错: {e}")在 rdf_queries.py 文件中使用的是 query_rdf 函数,其具体内容如下所示:
# 使用SPARQL查询RDF的函数
定义一个函数 query_rdf,参数包括:local_file_path, query, mesh_terms, 和可选参数 base_namespace,默认值为 'http://example.org/mesh/'。
def query_rdf(local_file_path, query, mesh_terms, base_namespace="http://example.org/mesh/"):
if not mesh_terms or not isinstance(mesh_terms, list):
raise ValueError("MeSH术语列表为空或格式不正确。")
print("SPARQL查询:", query)
# 创建并解析RDF图
g = Graph()
g.parse(local_file_path, format="ttl")
article_data = {}
for term in mesh_terms:
# 将术语转换为有效的URI
mesh_term_uri = convert_to_uri(term, base_namespace)
#print("术语:", term, "URI:", mesh_term_uri)
# 使用initBindings参数执行SPARQL查询
results = g.query(query, initBindings={'meshTerm': mesh_term_uri})
for row in results:
article_uri = row['article']
if article_uri not in article_data:
article_data[article_uri] = {
'title': row['title'],
'abstract': row['abstract'],
'日期发布': row['datePublished'],
'access': row['access'],
'meshTerms': set()
}
article_data[article_uri]['meshTerms'].add(str(row['meshTerm']))
#print("DEBUG article_data:", article_data)
# 根据与文章匹配的MeSH术语数量对文章进行排名
ranked_articles = sorted(
article_data.items(),
key=lambda item: len(item[1]['meshTerms']),
reverse=True
)
返回排名前10的文章正如你所见,这个功能还会把MeSH术语转换成URI,这样我们就可以利用图形来过滤。在把术语转成URI时,要小心处理,并确保这种转换方式与其他功能保持一致。
这就是应用界面。
这张图片是由作者提供的
正如您所见,我们从上一步中选择的两个MeSH术语在这里。如果我点击“筛选文章”,它将根据我们在第2步中设置的筛选条件来筛选最初的10篇文章。文章将返回其完整的摘要及其带标签的MeSH术语(请见下方图片)。
这张图片由作者制作
找到了5篇文章。其中两篇标为“嘴里的肿瘤”,一篇标为“牙龈上的肿瘤”,两篇标为“硬腭上的肿瘤”。
现在我们已经有了一个精简的文章列表,可以用来生成回复,我们可以进入最后一步。我们想要将这些文章发送给一个大型语言模型来生成回复,也可以在提示中加入额外的上下文信息。我有一个默认的提示,内容是:“用项目符号列出关键信息点。让没有医学背景的人也能理解。”在这个演示中,我会调整提示来反映最初的搜索词。
结果是这样的:
这些结果看起来更好,主要是因为据推测我们正在总结的文章大概都是关于口腔癌治疗的文章。数据集里没有实际的期刊文章,只有摘要。所以这些结果只是摘要的总结。这或许有它的价值,但如果我们是在开发一个真正的应用程序而不仅仅是演示,在这里,我们就可以加入文章的全文。或者用户/研究者会在这一环节自己去阅读这些文章,而不是完全依赖LLM提供的摘要。
结尾:总结一下本教程演示了结合向量数据库和知识图谱可以显著增强RAG应用。利用向量相似性进行初始搜索,并通过结构化的知识图谱元数据进行筛选和组织,我们可以构建一个能够提供准确、可解释且领域特定的结果的体系。集成MeSH这一广泛认可的受控词汇表,突显了这种优势,这确保检索步骤能够满足应用的独特需求,同时保持与其他系统的互操作性。这种方法不仅限于医学领域——其原则可以应用于任何结构化数据和文本信息共存的领域中。
本教程强调了利用每种技术的最佳优势的重要性。向量数据库在相似性检索方面表现出色,而知识图谱则在提供上下文、结构和语义方面更加出色。此外,扩展RAG应用程序需要一个元数据层来打破数据孤岛并实施治理政策,以确保数据的连贯性和合规性。周到的设计应基于特定领域的元数据和强大的治理,是构建既准确又可扩展的RAG系统的最佳路径。
, '', term)
# 第二步:将非单词字符替换为下划线(一个或多个)
formatted_term = re.sub(r'\W+', '_', stripped_term)
# 第三步:将连续的多个下划线替换为单个下划线
formatted_term = re.sub(r'_+', '_', formatted_term)
# 第四步:对术语进行URL编码,处理任何剩余的特殊字符
encoded_term = quote(formatted_term)
# 第五步:添加单个首尾下划线
term_with_underscores = f"_{encoded_term}_"
# 第六步:连接命名空间和术语,不添加额外的下划线
uri = f"{base_namespace}{term_with_underscores}"
return URIRef(uri)
# 生成过去5年内随机日期
def generate_random_date():
start_date = datetime.now() - timedelta(days=5*365)
random_days = random.randint(0, 5*365)
return start_date + timedelta(days=random_days)
# 生成随机访问值 (1到10之间)
def generate_random_access():
return random.randint(1, 10)
# 通过替换非单词字符为下划线并进行URL编码来生成文章的URI
def create_article_uri(title, base_namespace="http://example.org/article"):
"""
参数:
title (str): 文章的标题。
base_namespace (str): 文章URI的基础命名空间。
返回:
URIRef: 格式化的文章URI。
"""
if pd.isna(title):
return None
# 对文本进行编码以用于URI
sanitized_text = urllib.parse.quote(title.strip().replace(' ', '_').replace('"', '').replace('<', '').replace('>', '').replace("'", "_"))
return URIRef(f"{base_namespace}/{sanitized_text}")
# 遍历DataFrame中的每一行以创建RDF三元组
for index, row in df.iterrows():
article_uri = create_article_uri(row['Title'])
if article_uri is None:
continue
# 添加Article实例
g.add((article_uri, RDF.type, Article))
g.add((article_uri, title, Literal(row['Title'], datatype=XSD.string)))
g.add((article_uri, abstract, Literal(row['abstractText'], datatype=XSD.string)))
# 添加随机datePublished和access
random_date = generate_random_date()
random_access = generate_random_access()
g.add((article_uri, date_published, Literal(random_date.date(), datatype=XSD.date)))
g.add((article_uri, access, Literal(random_access, datatype=XSD.integer)))
# 添加MeSH Terms
mesh_terms = parse_mesh_terms(row['meshMajor'])
for term in mesh_terms:
term_uri = convert_to_uri(term, base_namespace="http://example.org/mesh/")
if term_uri is None:
continue
# 添加MeSH Term实例
g.add((term_uri, RDF.type, MeSHTerm))
g.add((term_uri, RDFS.label, Literal(term.replace('_', ' '), datatype=XSD.string)))
# 将Article链接到MeSH Term
g.add((article_uri, schema.about, term_uri))
# 文件保存路径
file_path = "/Workspace/PubMedGraph.ttl"
# 保存文件
g.serialize(destination=file_path, format='turtle')
print(f"文件已保存至 {file_path}")
OK, so now we have a vectorized version of the data, and a graph (RDF) version of the data. Each vector has a URI associated with it, which corresponds to an entity in the KG, so we can go back and forth between the data formats.
# Build an app
I decided to use [Streamlit](https://streamlit.io/) to build the interface for this graph RAG app. Similar to the last blog post, I have kept the user flow the same.
1. **Search Articles:** First, the user searches for articles using a search term. This relies exclusively on the vector database. The user’s search term(s) is sent to the vector database and the ten articles nearest the term in vector space are returned.
2. **Refine Terms:** Second, the user decides the MeSH terms to use to filter the returned results. Since we also vectorized the MeSH terms, we can have the user enter a natural language prompt to get the most relevant MeSH terms. Then, we allow the user to expand these terms to see their alternative names and narrower concepts. The user can select as many terms as they want for their filter criteria.
3. **Filter & Summarize: **Third, the user applies the selected terms as filters to the original ten journal articles. We can do this since the PubMed articles are tagged with MeSH terms. Finally, we let the user enter an additional prompt to send to the LLM along with the filtered journal articles. This is the generative step of the RAG app.
Let’s go through these steps one at a time. You can see the full app and code on my GitHub, but here is the structure:
-- app.py (a python file that drives the app and calls other functions as needed)
-- query_functions (a folder containing python files with queries)
-- rdf_queries.py (python file with RDF queries)
-- weaviate_queries.py (python file containing weaviate queries)
-- PubMedGraph.ttl (the pubmed data in RDF format, stored as a ttl file)
## Search Articles
First, want to do is implement Weaviate’s [vector similarity search](https://weaviate.io/developers/weaviate/search/similarity). Since our articles are vectorized, we can send a search term to the vector database and get similar articles back.
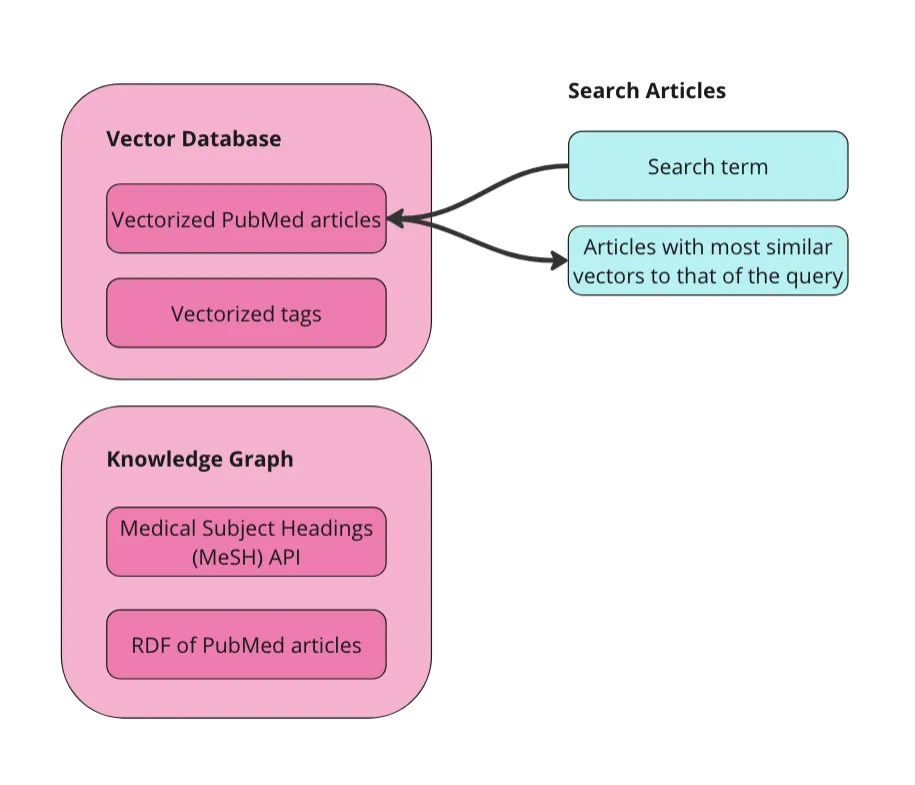
Image by Author
The main function that searches for relevant journal articles in the vector database is in app.py:
# --- TAB 1: Search Articles ---
with tab_search:
st.header("Search Articles (Vector Query)")
query_text = st.text_input("Enter your vector search term (e.g., Mouth Neoplasms):", key="vector_search")
if st.button("Search Articles", key="search_articles_btn"):
try:
client = initialize_weaviate_client()
article_results = query_weaviate_articles(client, query_text)
# Extract URIs here
article_uris = [
result["properties"].get("article_URI")
for result in article_results
if result["properties"].get("article_URI")
]
# Store article_uris in the session state
st.session_state.article_uris = article_uris
st.session_state.article_results = [
{
"Title": result["properties"].get("title", "N/A"),
"Abstract": (result["properties"].get("abstractText", "N/A")[:100] + "..."),
"Distance": result["distance"],
"MeSH Terms": ", ".join(
ast.literal_eval(result["properties"].get("meshMajor", "[]"))
if result["properties"].get("meshMajor") else []
),
}
for result in article_results
]
client.close()
except Exception as e:
st.error(f"Error during article search: {e}")
if st.session_state.article_results:
st.write("**Search Results for Articles:**")
st.table(st.session_state.article_results)
else:
st.write("No articles found yet.")
This function uses the queries stored in weaviate_queries to establish the Weaviate client (initialize_weaviate_client) and search for articles (query_weaviate_articles). Then we display the returned articles in a table, along with their abstracts, distance (how close they are to the search term), and the MeSH terms that they are tagged with.
The function to query Weaviate in weaviate_queries.py looks like this:
# Function to query Weaviate for Articles
def query_weaviate_articles(client, query_text, limit=10):
# Perform vector search on Article collection
response = client.collections.get("Article").query.near_text(
query=query_text,
limit=limit,
return_metadata=MetadataQuery(distance=True)
)
# Parse response
results = []
for obj in response.objects:
results.append({
"uuid": obj.uuid,
"properties": obj.properties,
"distance": obj.metadata.distance,
})
return results
As you can see, I put a limit of ten results here just to make it simpler, but you can change that. This is just using vector similarity search in Weaviate to return relevant results.
The end result in the app looks like this:
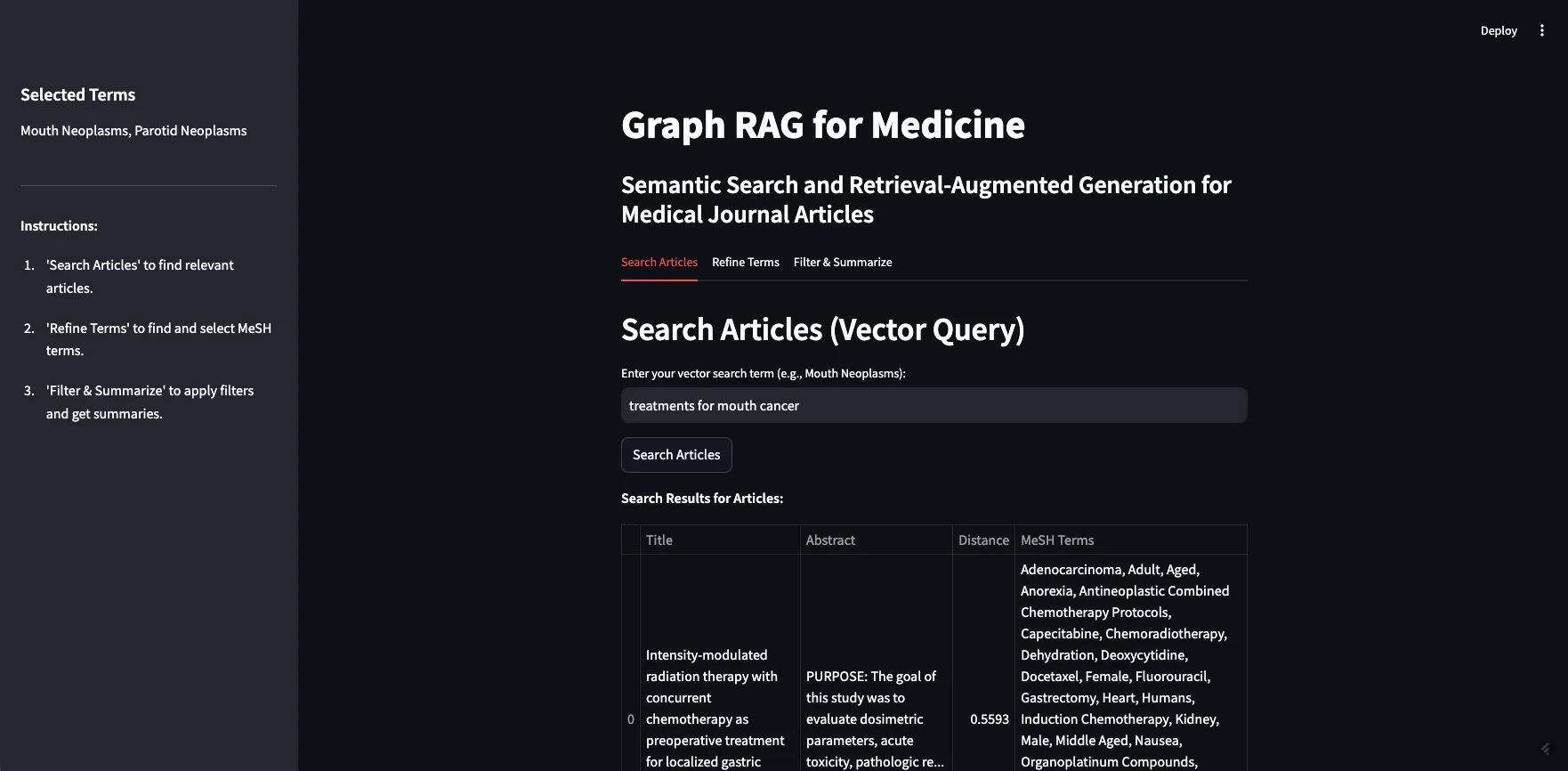
Image by Author
As a demo, I will search the term “treatments for mouth cancer”. As you can see, 10 articles are returned, mostly relevant. This demonstrates both the strengths and weaknesses of vector based retrieval.
The strength is that we can build a semantic search functionality on our data with minimal effort. As you can see above, all we did was set up the client and send the data to a vector database. Once our data has been vectorized, we can do semantic searches, similarity searches, and even RAG. I have put some of that in the notebook accompanying this post, but there’s a lot more in Weaviate’s [official docs](https://weaviate.io/developers/weaviate).
The weakness of vector based retrieval, as I mentioned above are that they are black-box and struggle with factual knowledge. In our example, it looks like most of the articles are about some kind of treatment or therapy for some kind of cancer. Some of the articles are about mouth cancer specifically, some are about a sub-type of mouth cancer like gingival cancer (cancer of the gums), and palatal cancer (cancer of the palate). But there are also articles about nasopharyngeal cancer (cancer of the upper throat), mandibular cancer (cancer of the jaw), and esophageal cancer (cancer of the esophagus). None of these (upper throat, jaw, or esophagus) are considered mouth cancer. It is understandable why an article about a specific cancer radiation therapy for nasopharyngeal neoplasms would be considered similar to the prompt “treatments for mouth cancer” but it may not be relevant if you are only looking for treatments for mouth cancer. If we were to plug these ten articles directly into our prompt to the LLM and ask it to “summarize the different treatment options,” we would be getting incorrect information.
The purpose of RAG is to give an LLM a very specific set of additional information to better answer your question — if that information is incorrect or irrelevant, it can lead to misleading responses from the LLM. This is often referred to as “context poisoning”. What is especially dangerous about context poisoning is that the response isn’t necessarily factually inaccurate (the LLM may accurately summarize the treatment options we feed it), and it isn’t necessarily based on an inaccurate piece of data (presumably the journal articles themselves are accurate), it’s just using the wrong data to answer your question. In this example, the user could be reading about how to treat the wrong kind of cancer, which seems very bad.
## Refine Terms
KGs can help improve the accuracy of responses and reduce the likelihood of context poisoning by refining the results from the vector database. The next step is for selecting what MeSH terms we want to use to filter the articles. First, we do another vector similarity search against the vector database but on the Terms collection. This is because the user may not be familiar with the MeSH controlled vocabulary. In our example above, I searched for, “therapies for mouth cancer”, but “mouth cancer” is not a term in MeSH — they use “Mouth Neoplasms”. We want the user to be able to start exploring the MeSH terms without having a prior understanding of them — this is good practice regardless of the metadata used to tag the content.
Image by Author
The function to get relevant MeSH terms is nearly identical to the previous Weaviate query. Just replace Article with term:
# Function to query Weaviate for MeSH Terms
def query_weaviate_terms(client, query_text, limit=10):
# Perform vector search on MeshTerm collection
response = client.collections.get("term").query.near_text(
query=query_text,
limit=limit,
return_metadata=MetadataQuery(distance=True)
)
# Parse response
results = []
for obj in response.objects:
results.append({
"uuid": obj.uuid,
"properties": obj.properties,
"distance": obj.metadata.distance,
})
return results
Here is what it looks like in the app:
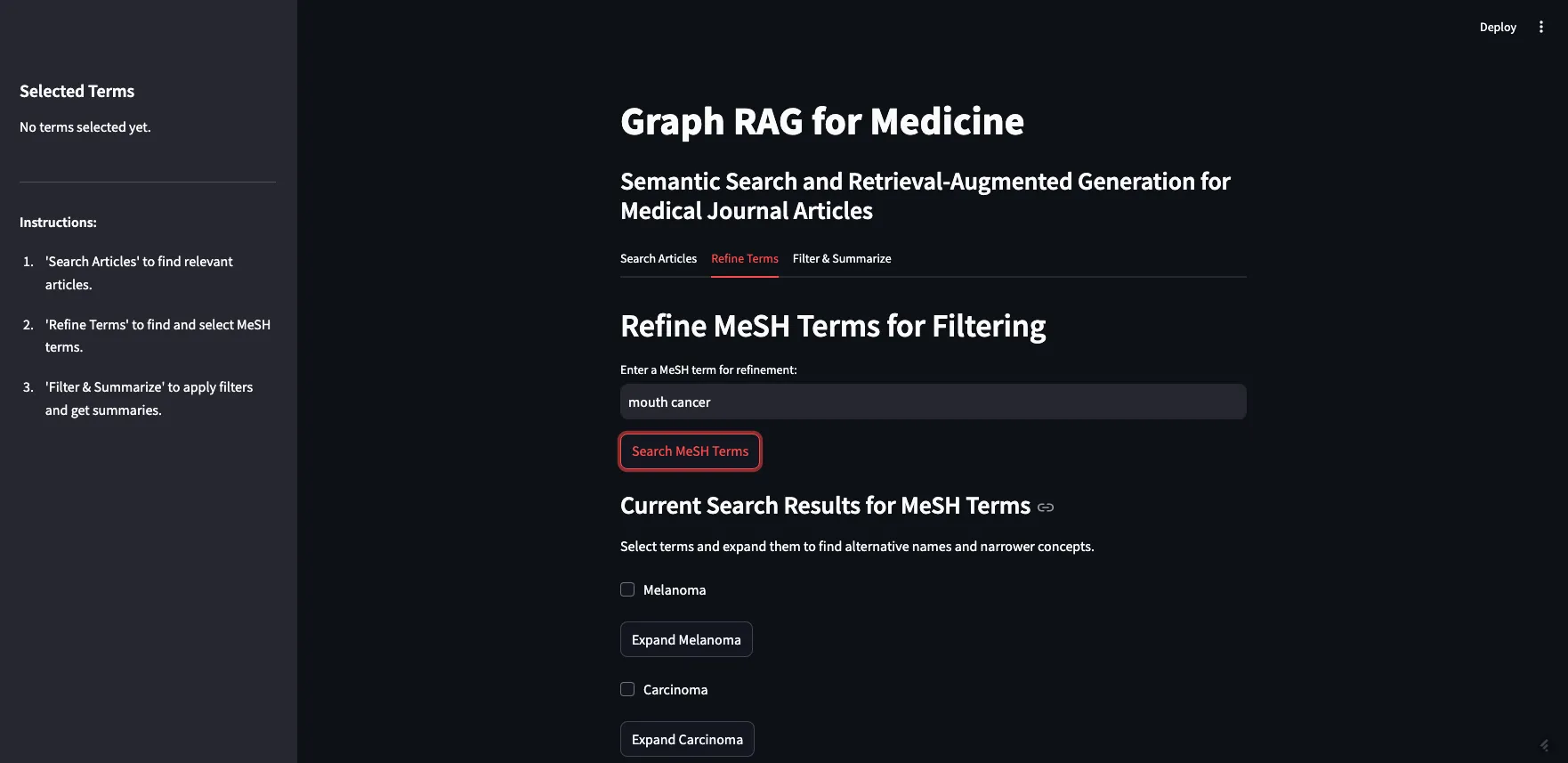
Image by Author
As you can see, I searched for “mouth cancer” and the most similar terms were returned. Mouth cancer was not returned, as that is not a term in MeSH, but Mouth Neoplasms is on the list.
The next step is to allow the user to expand the returned terms to see alternative names and narrower concepts. This requires querying the [MeSH API](https://id.nlm.nih.gov/mesh/). This was the trickiest part of this app for a number of reasons. The biggest problem is that Streamlit requires that everything has a unique ID but MeSH terms can repeat — if one of the returned concepts is a child of another, then when you expand the parent you will have a duplicate of the child. I think I took care of most of the big issues and the app should work, but there are probably bugs to find at this stage.
The functions we rely on are found in rdf_queries.py. We need one to get the alternative names for a term:
# Fetch alternative names and triples for a MeSH term
def get_concept_triples_for_term(term):
term = sanitize_term(term) # Sanitize input term
sparql = SPARQLWrapper("https://id.nlm.nih.gov/mesh/sparql")
query = f"""
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX meshv: <http://id.nlm.nih.gov/mesh/vocab#>
PREFIX mesh: <http://id.nlm.nih.gov/mesh/>
SELECT ?subject ?p ?pLabel ?o ?oLabel
FROM <http://id.nlm.nih.gov/mesh>
WHERE {{
?subject rdfs:label "{term}"@en .
?subject ?p ?o .
FILTER(CONTAINS(STR(?p), "concept"))
OPTIONAL {{ ?p rdfs:label ?pLabel . }}
OPTIONAL {{ ?o rdfs:label ?oLabel . }}
}}
"""
try:
sparql.setQuery(query)
sparql.setReturnFormat(JSON)
results = sparql.query().convert()
triples = set()
for result in results["results"]["bindings"]:
obj_label = result.get("oLabel", {}).get("value", "No label")
triples.add(sanitize_term(obj_label)) # Sanitize term before adding
# Add the sanitized term itself to ensure it's included
triples.add(sanitize_term(term))
return list(triples)
except Exception as e:
print(f"Error fetching concept triples for term '{term}': {e}")
return []
We also need functions to get the narrower (child) concepts for a given term. I have two functions that achieve this — one that gets the immediate children of a term and one recursive function that returns all children of a given depth.
# Fetch narrower concepts for a MeSH term
def get_narrower_concepts_for_term(term):
term = sanitize_term(term) # Sanitize input term
sparql = SPARQLWrapper("https://id.nlm.nih.gov/mesh/sparql")
query = f"""
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX meshv: <http://id.nlm.nih.gov/mesh/vocab#>
PREFIX mesh: <http://id.nlm.nih.gov/mesh/>
SELECT ?narrowerConcept ?narrowerConceptLabel
WHERE {{
?broaderConcept rdfs:label "{term}"@en .
?narrowerConcept meshv:broaderDescriptor ?broaderConcept .
?narrowerConcept rdfs:label ?narrowerConceptLabel .
}}
"""
try:
sparql.setQuery(query)
sparql.setReturnFormat(JSON)
results = sparql.query().convert()
concepts = set()
for result in results["results"]["bindings"]:
subject_label = result.get("narrowerConceptLabel", {}).get("value", "No label")
concepts.add(sanitize_term(subject_label)) # Sanitize term before adding
return list(concepts)
except Exception as e:
print(f"Error fetching narrower concepts for term '{term}': {e}")
return []
# Recursive function to fetch narrower concepts to a given depth
def get_all_narrower_concepts(term, depth=2, current_depth=1):
term = sanitize_term(term) # Sanitize input term
all_concepts = {}
try:
narrower_concepts = get_narrower_concepts_for_term(term)
all_concepts[sanitize_term(term)] = narrower_concepts
if current_depth < depth:
for concept in narrower_concepts:
child_concepts = get_all_narrower_concepts(concept, depth, current_depth + 1)
all_concepts.update(child_concepts)
except Exception as e:
print(f"Error fetching all narrower concepts for term '{term}': {e}")
return all_concepts
The other important part of step 2 is to allow the user to select terms to add to a list of “Selected Terms”. These will appear in the sidebar on the left of the screen. There are a lot of things that can improve this step like:
* There is no way to clear all but you can clear the cache or refresh the browser if needed.
* There is no way to ‘select all narrower concepts’ which would be helpful.
* There is no option to add rules for filtering. Right now, we are just assuming that the article must contain term A OR term B OR term C etc. The rankings at the end are based on the number of terms the articles are tagged with.
Here is what it looks like in the app:
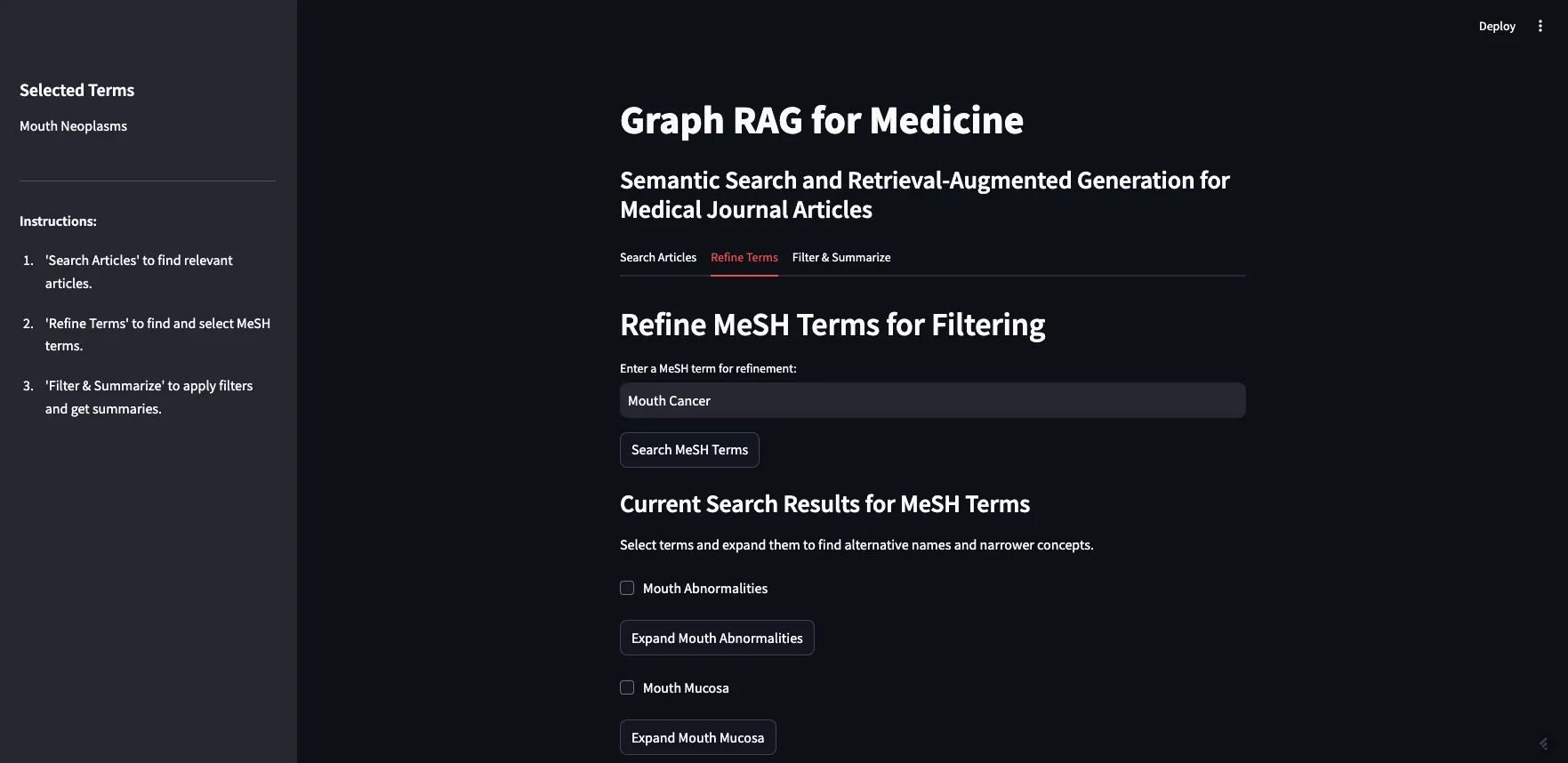
Image by Author
I can expand Mouth Neoplasms to see the alternative names, in this case, “Cancer of Mouth”, along with all of the narrower concepts. As you can see, most of the narrower concepts have their own children, which you can expand as well. For the purposes of this demo, I am going to select all children of Mouth Neoplasms.
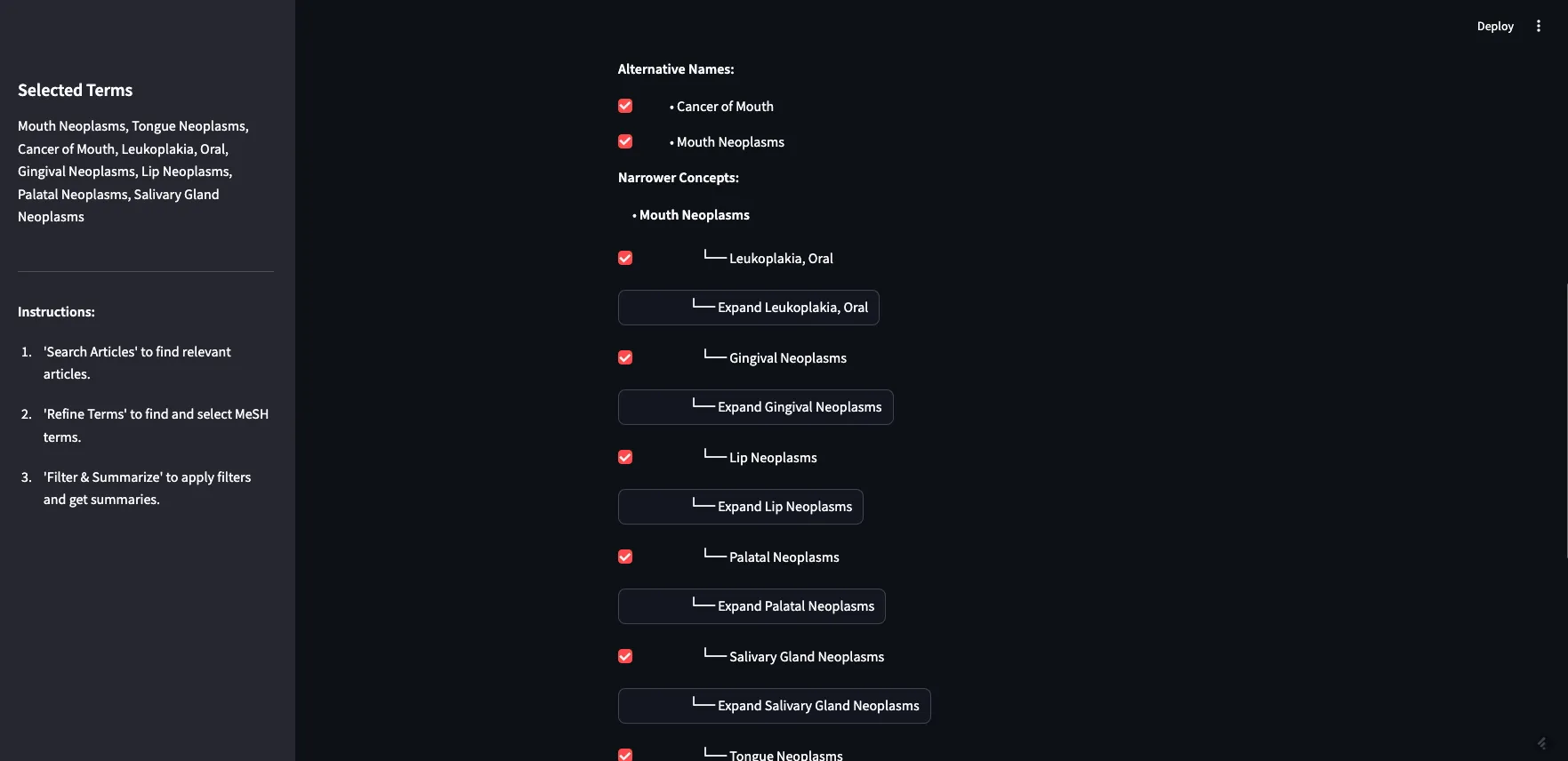
Image by Author
This step is important not just because it allows the user to filter the search results, but also because it is a way for the user to explore the MeSH graph itself and learn from it. For example, this would be the place for the user to learn that nasopharyngeal neoplasms are not a subset of mouth neoplasms.
## Filter & Summarize
Now that you’ve got your articles and your filter terms, you can apply the filter and summarize the results. This is where we bring the original 10 articles returned in step one together with the refined list of MeSH terms. We allow the user to add additional context to the prompt before sending it to the LLM.
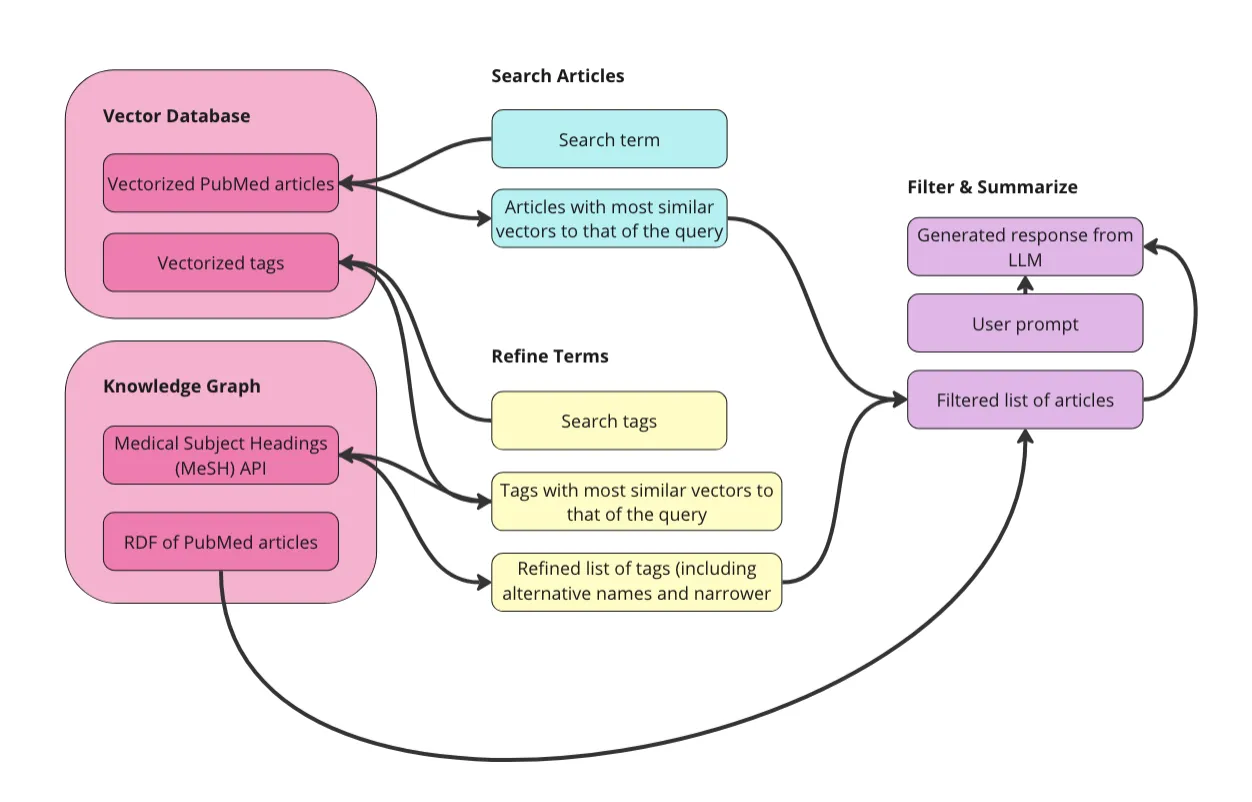
Image by Author
The way we do this filtering is that we need to get the URIs for the 10 articles from the original search. Then we can query our knowledge graph for which of those articles have been tagged with the associated MeSH terms. Additionally, we save the abstracts of these articles for use in the next step. This would be the place where we could filter based on access control or other user-controlled parameters like author, filetype, date published, etc. I didn’t include any of that in this app but I did add in properties for access control and date published in case we want to add that in this UI later.
Here is what the code looks like in app.py:
if st.button("Filter Articles"):
try:
# Check if we have URIs from tab 1
if "article_uris" in st.session_state and st.session_state.article_uris:
article_uris = st.session_state.article_uris
# Convert list of URIs into a string for the VALUES clause or FILTER
article_uris_string = ", ".join([f"<{str(uri)}>" for uri in article_uris])
SPARQL_QUERY = """
PREFIX schema: <http://schema.org/>
PREFIX ex: <http://example.org/>
SELECT ?article ?title ?abstract ?datePublished ?access ?meshTerm
WHERE {{
?article a ex:Article ;
schema:name ?title ;
schema:description ?abstract ;
schema:datePublished ?datePublished ;
ex:access ?access ;
schema:about ?meshTerm .
?meshTerm a ex:MeSHTerm .
FILTER (?article IN ({article_uris}))
}}
"""
# Insert the article URIs into the query
query = SPARQL_QUERY.format(article_uris=article_uris_string)
else:
st.write("No articles selected from Tab 1.")
st.stop()
# Query the RDF and save results in session state
top_articles = query_rdf(LOCAL_FILE_PATH, query, final_terms)
st.session_state.filtered_articles = top_articles
if top_articles:
# Combine abstracts from top articles and save in session state
def combine_abstracts(ranked_articles):
combined_text = " ".join(
[f"Title: {data['title']} Abstract: {data['abstract']}" for article_uri, data in
ranked_articles]
)
return combined_text
st.session_state.combined_text = combine_abstracts(top_articles)
else:
st.write("No articles found for the selected terms.")
except Exception as e:
st.error(f"Error filtering articles: {e}")
This uses the function query_rdf in the rdf_queries.py file. That function looks like this:
# Function to query RDF using SPARQL
def query_rdf(local_file_path, query, mesh_terms, base_namespace="http://example.org/mesh/"):
if not mesh_terms:
raise ValueError("The list of MeSH terms is empty or invalid.")
print("SPARQL Query:", query)
# Create and parse the RDF graph
g = Graph()
g.parse(local_file_path, format="ttl")
article_data = {}
for term in mesh_terms:
# Convert the term to a valid URI
mesh_term_uri = convert_to_uri(term, base_namespace)
#print("Term:", term, "URI:", mesh_term_uri)
# Perform SPARQL query with initBindings
results = g.query(query, initBindings={'meshTerm': mesh_term_uri})
for row in results:
article_uri = row['article']
if article_uri not in article_data:
article_data[article_uri] = {
'title': row['title'],
'abstract': row['abstract'],
'datePublished': row['datePublished'],
'access': row['access'],
'meshTerms': set()
}
article_data[article_uri]['meshTerms'].add(str(row['meshTerm']))
#print("DEBUG article_data:", article_data)
# Rank articles by the number of matching MeSH terms
ranked_articles = sorted(
article_data.items(),
key=lambda item: len(item[1]['meshTerms']),
reverse=True
)
return ranked_articles[:10]
As you can see, this function also converts the MeSH terms to URIs so we can filter using the graph. Be careful in the way you convert terms to URIs and ensure it aligns with the other functions.
Here is what it looks like in the app:
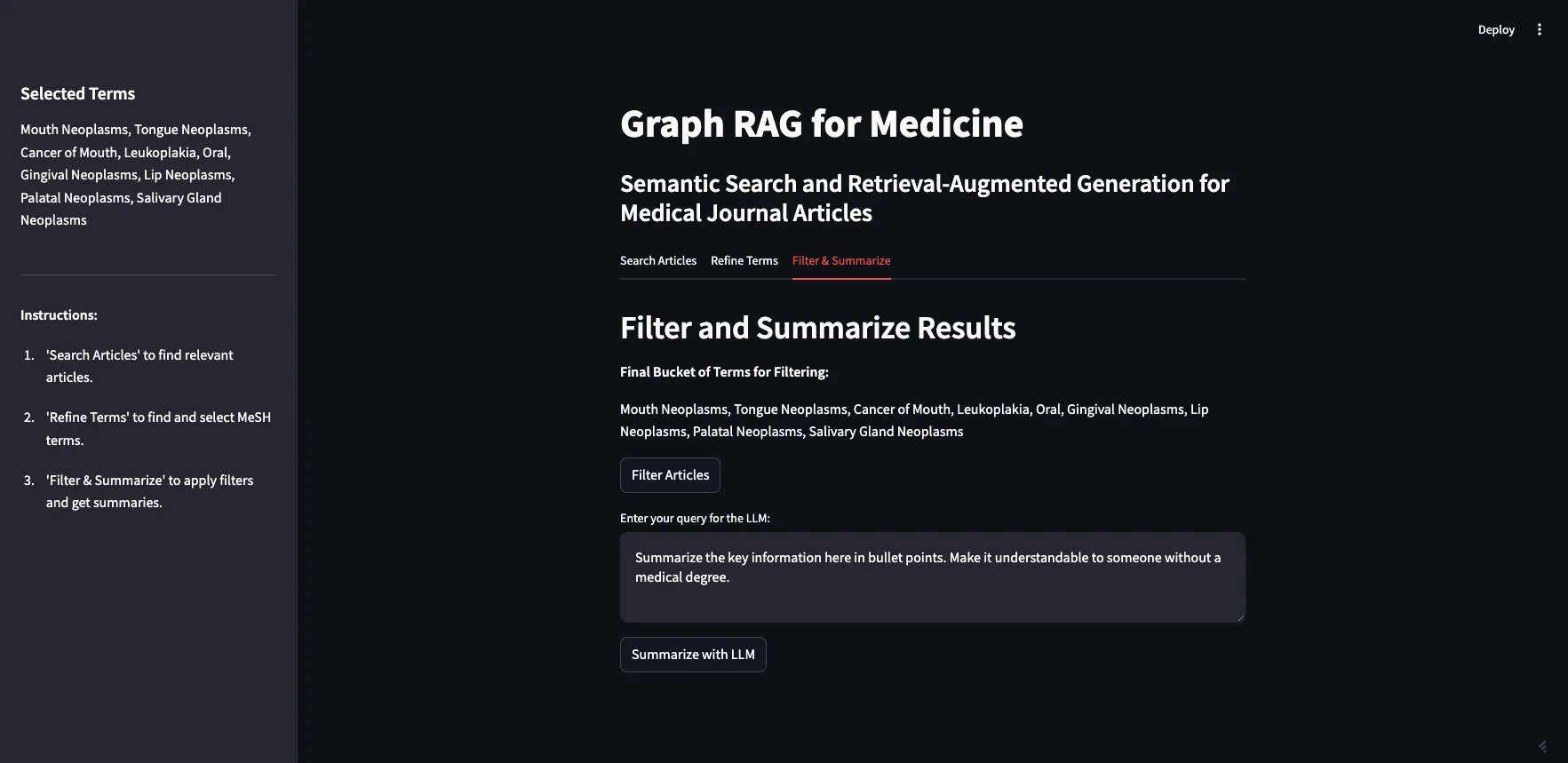
Image by Author
As you can see, the two MeSH terms we selected from the previous step are here. If I click “Filter Articles,” it will filter the original 10 articles using our filter criteria in step 2. The articles will be returned with their full abstracts, along with their tagged MeSH terms (see image below).
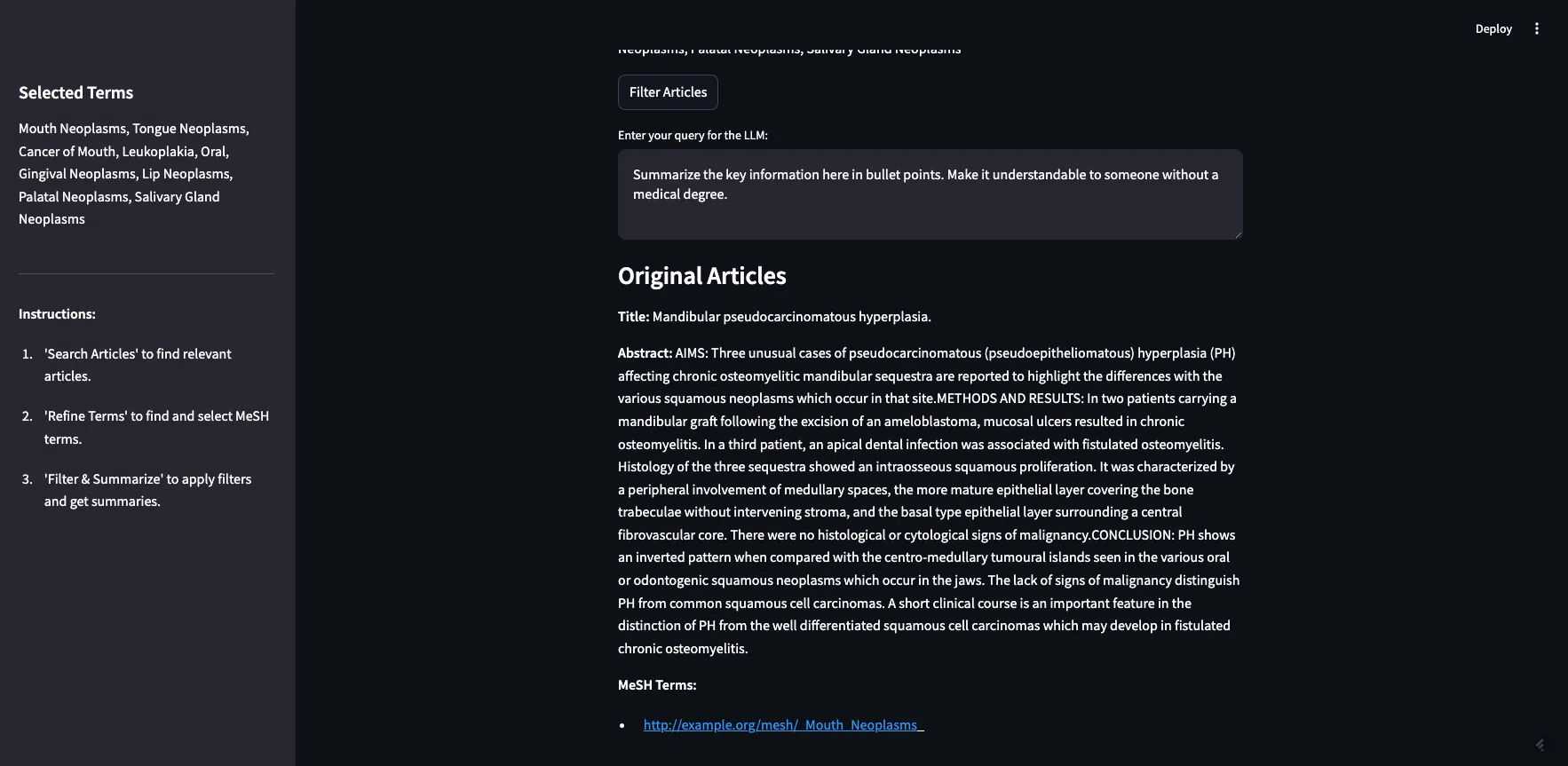
Image by Author
There are 5 articles returned. Two are tagged with “mouth neoplasms,” one with “gingival neoplasms,” and two with “palatal neoplasms”.
Now that we have a refined list of articles we want to use to generate a response, we can move to the final step. We want to send these articles to an LLM to generate a response but we can also add in additional context to the prompt. I have a default prompt that says, “Summarize the key information here in bullet points. Make it understandable to someone without a medical degree.” For this demo, I am going to adjust the prompt to reflect our original search term:
The results are as follows:

The results look better to me, mostly because I know that the articles we are summarizing are, presumably, about treatments for mouth cancer. The dataset doesn’t contain the actual journal articles, just the abstracts. So these results are just summaries of summaries. There may be some value to this, but if we were building a real app and not just a demo, this is the step where we could incorporate the full text of the articles. Alternatively, this is where the user/researcher would go read these articles themselves, rather than relying exclusively on the LLM for the summaries.
# Conclusion
This tutorial demonstrates how combining vector databases and knowledge graphs can significantly enhance RAG applications. By leveraging vector similarity for initial searches and structured knowledge graph metadata for filtering and organization, we can build a system that delivers accurate, explainable, and domain-specific results. The integration of MeSH, a well-established controlled vocabulary, highlights the power of domain expertise in curating metadata, which ensures that the retrieval step aligns with the unique needs of the application while maintaining interoperability with other systems. This approach is not limited to medicine — its principles can be applied across domains wherever structured data and textual information coexist.
This tutorial underscores the importance of leveraging each technology for what it does best. Vector databases excel at similarity-based retrieval, while knowledge graphs shine in providing context, structure, and semantics. Additionally, scaling RAG applications demands a metadata layer to break down data silos and enforce governance policies. Thoughtful design, rooted in domain-specific metadata and robust governance, is the path to building RAG systems that are not only accurate but also scalable.共同学习,写下你的评论
评论加载中...
作者其他优质文章